SVD, pseudoinverse, and PCA: SVD, pseudoinverse and PCA in Python
 PseudoInverse
PseudoInverse
The Python package numpy.linalg contains the function pinv to determine numerically the pseudoinverse of a matrix.
Example 1: linear regression First we calculate the model from the lesson text as follows:
>>> import numpy as np
>>> import numpy.linalg as la
>>> A = np.array([[1,1],[1,2],[1,3]]); print(A) # matrix A for regression line
[[1 1]
[1 2]
[1 3]]
>>> b = np.array([2,2,4]); print(b) # data vector
[2 2 4]
>>> Ainv = la.pinv(A); print(np.round(Ainv, 4)) # pseudoinverse
[[ 1.3333 0.3333 -0.6667]
[-0.5 0. 0.5 ]]
>>> print(np.round(Ainv @ b, 4)) # regression coefficients
[0.6667 1. ] Actually, the result is in front of your eyes here: \(y= 0.6667+ t\). We can also calculate this outcome via singular value decomposition.
>>> U, S, Vt = la.svd(A, full_matrices=False) # SVD van A
>>> print(U) # matrix U in SVD
[[-0.32310986 0.85377594]
[-0.54750702 0.18321952]
[-0.77190419 -0.4873369 ]]
>>> print(np.diag(S)) # diagonal matrix with singular values
[[4.07914333 0. ]
[0. 0.60049122]]
>>> print(Vt) # transposed of matrix V in SVD
[[-0.40266324 -0.91534819]
[ 0.91534819 -0.40266324]]
>>> print(U @ np.diag(S) @ Vt) # check of SVD calculation
[[1., 1.],
[1., 2.],
[1., 3.]])
>>> Splus = la.inv(np.diag(S)); print(Splus)
[[0.24514951, 0. ]
[0. , 1.66530329]
>>> Ainv = Vt.transpose() @ Splus @ U.transpose()
>>> print(np.round(Ainv, 4))
[[ 1.3333 0.3333 -0.6667]
[-0.5 0. 0.5 ]]
>>> print(np.round(Ainv @ b, 4)) # regression coefficients
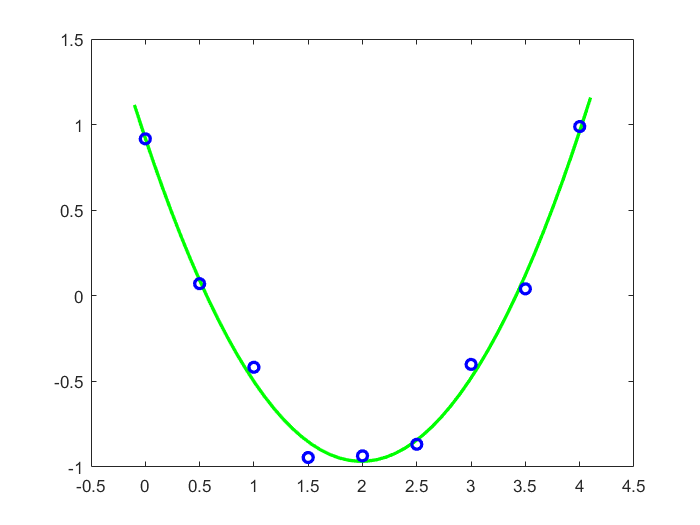
[0.6667 1. ] Example 2: quadratic regression The second example is a data set that we create by adding noise to the parabola \(1-2x+\frac{1}{2}x^2\). Hereafter we search for the parabola that fits the created data set best. In our data set this is approximately \(y=0.98-1.94x+0.49x^2\).
>>> import numpy as np
>>> import numpy.linalg as la
>>> import matplotlib.pyplot as plt
>>> x = np.arange(0, 4, 0.5)
>>> y = 1 - 2*x + 1/2*x**2 # parabola
>>> y = y + 0.1*max(y)*(2*np.random.randint(2, size=m) - np.ones(m)) # 10% noise added
>>> A = np.vstack((np.ones(m), x, x**2)).transpose()
>>> pinvA = la.pinv(A)
>>> c = pinvA @ y; print(c) # pseudoinverse of A gives regression coefficients
[ 0.975 -1.93571429 0.48571429]
>>> xx = np.linspace(-0.1,4.1,50)
>>> yy = c[0] + c[1]*xx + c[2]*xx**2
>>> plt.plot(x, y, 'bo')
>>> plt.plot(xx, yy, 'g-')
>>> plt.axis([-0.1, 4.1, -1.1, 1.1])
>>> plt.show() # visualisation of data + approximation
Unlock full access



