Bioelectricity: Electric excitability and action potential
 Simulation of the Hodgkin-Huxley model in R
Simulation of the Hodgkin-Huxley model in R
The model of Hodgkin and Huxley can be simulated in a mathematical software environment, for example in R and RStudio.
The R script below simulates the model for a momentary stimulation high enough to trigger an action potential. For clarity, the shifted electrical potential \(U=V-V_r\) is modeled here with resting membrane potential \(V_r\), and \(V=U+V_r\) is calculated after solving the system of differential equations.
The R script below simulates the model for a momentary stimulation high enough to trigger an action potential. For clarity, the shifted electrical potential \(U=V-V_r\) is modeled here with resting membrane potential \(V_r\), and \(V=U+V_r\) is calculated after solving the system of differential equations.
library(deSolve)
model <- function(tijd, begintoestand, parameters) {
with(as.list(c(begintoestand, parameters)),{
an <- function(U) 0.01*(10-U)/(exp(1-0.1*U)-1);
bn <- function(U) 0.125*exp(-0.0125*U);
am <- function(U) (2.5-0.1*U)/(exp(2.5-0.1*U)-1);
bm <- function(U) 4*exp(-0.0556*U);
ah <- function(U) 0.07*exp(-0.05*U);
bh <- function(U) 1/(exp(3-0.1*U)+1);
Istim <- function(t, s=start, d=duur) {
if (t>s && t<s+d) {I} else {0}
} # stroomstimulus in blokvorm met hoogste waarde I
dUdt <- (Istim(tijd) - gk*n^4*(U-Uk) - gna*m^3*h*(U-Una) - gl*(U-Ul))/C;
dndt <- an(U)*(1-n)-bn(U)*n;
dmdt <- am(U)*(1-m)-bm(U)*m;
dhdt <- ah(U)*(1-h)-bh(U)*h;
return(list(c(dUdt, dndt, dmdt, dhdt)))
})
}
params <- c(Una=115, Uk=-12, Ul=10.6,
gna=120, gk=36, gl=0.3, C=1,
I=10, start=5, duur=1);
t <- seq(from=0, to=30, by = 0.1);
beginwaarden <- c(U=0, n=0.32, m=0.06, h=0.6);
oplossing <- ode(beginwaarden, t, model, params);
oplossing[,2] <- oplossing[,2] - 70 # V=U+Vrust; Vrust=-70 mV
plot(oplossing, col="blue", lwd=3,
xlab="tijd (msec)", ylab=list("V (mV)", "n", "m", "h"),
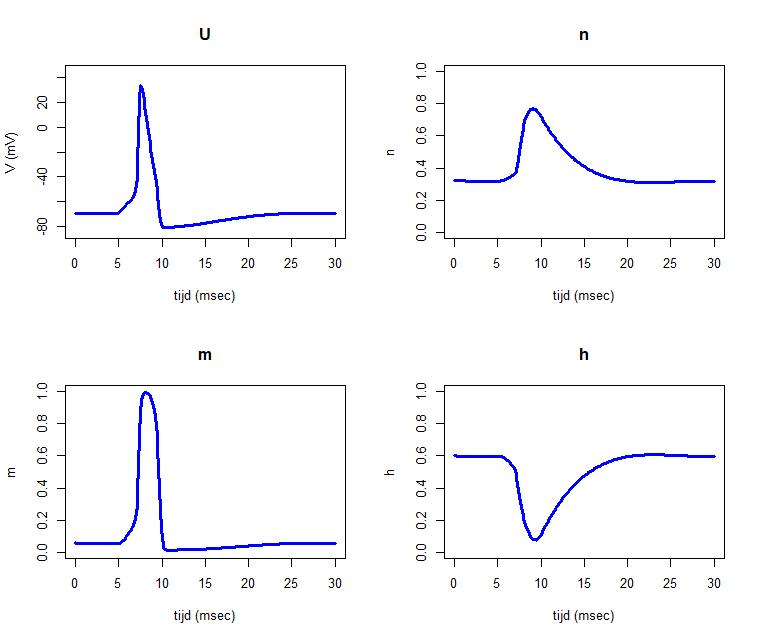
ylim=list(c(-85,45), c(0,1), c(0,1), c(0,1)))It produces the diagrams below.
Unlock full access



