Doing mathematics with R: Regression analysis in R
 Nonlinear regression via the peeling-off method
Nonlinear regression via the peeling-off method
The peeling-off method In the case of a bi-exponential model or a regression model composed of sums of sinusoids, applying a nonlinear regression method in one go may not be successful unless you already have good initial parameter values. In such a situation, the peeling-off method can be helpful. You start with an approximation using one generalised exponential model or one sinusoid. Then, you calculate the residuals and apply the same regression model to them. The sum of the two obtained regression curves serves as an initial approximation to the original nonlinear model and provides suitable starting values for an iterative method.
Two examples make this clearer (click on the tabs below).
Bi-exponential model of the head of a beer Every beer drinker is familiar with the phenomenon: when beer is poured into a glass, a head of the beer forms that disappears after a while if the glass remains untouched. The decreasing height of the head of a beer can be mathematically modelled reasonably well with a bi-exponential model. This approximation can be determined as follows (data file in csv format). The R script is executed and explained below in steps.
Step 1: reading in and plotting measured data
# read data from the csv file (measurements every 2 seconds)
data <- read.csv("beerfoam.csv", header = TRUE,
sep = ";", dec = ".", na.strings="", fill = TRUE)
t <- data$time # time in sec
H <- data$height # height of foam in cm
# plot measured data in a scatter plot
plot(t, H, type="p", pch=1, cex=0.7, col="red",
xlab="t (in sec)", ylab="height (in cm)",
main="Height of foam versus time")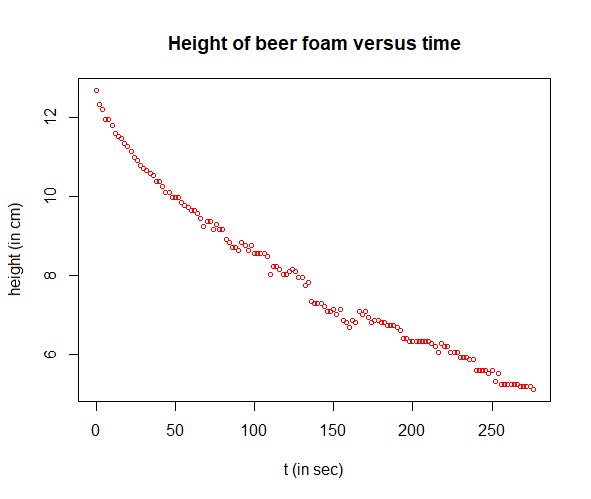
Step 2: describing the second part of measured data with exponential regression
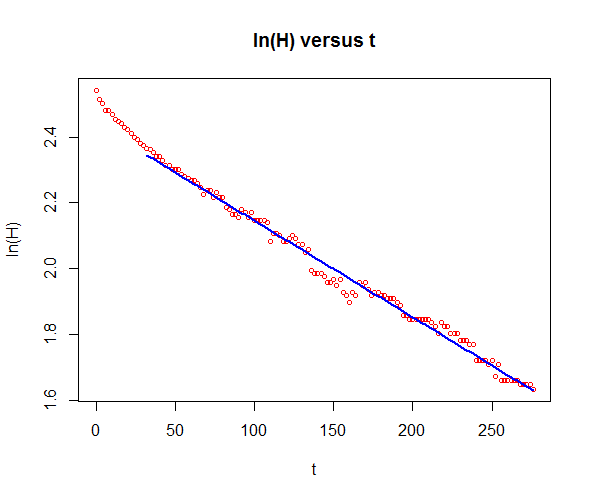
If you find an exponential growth model plausible, you can verify it by plotting the logarithm of the quantity against time as this should yield a straight line. In this specific case you see that over time, say when \(t>30\), a straight line does indeed describe the logarithm of the height well. You can then apply the exponential growth model to this selected piece.
The R script below produces the non-linear function fit and a matching diagram.
# plot ln(H.data) against t.data
plot(t, log(H), type="p", pch=1, cex=0.7, col="red",
xlab="t", ylab="ln(H)",
main="ln(H) versus t")
# select data after 30 seconds
dataselection <- c(t.data>30)
ts <- t[dataselection]
Hs <- H[dataselection]
lnHs <- log(Hs)
# linear regression of ln(Hs) as linear function of t
fit <- lm(formula = lnHs ~ ts)
lines(ts, predict(fit), type="l", col="blue", lwd=2)
Next, we plot the measured data and the exponential growth model.
# plot measured data in a scatter plot and add the expontial fit
coeffs <- coefficients(fit)
a1 <- exp(coeffs[1])
r1 <- coeffs[2]
H_fit <- a1*exp(r1*t)
plot(t, H, type="p", pch=1, cex=0.7, col="red",
xlab="t (in sec)", ylab="height (in cm)",
main="Height versus time with exponential model")
lines(t, H_fit, type="l", col="blue", lwd=2)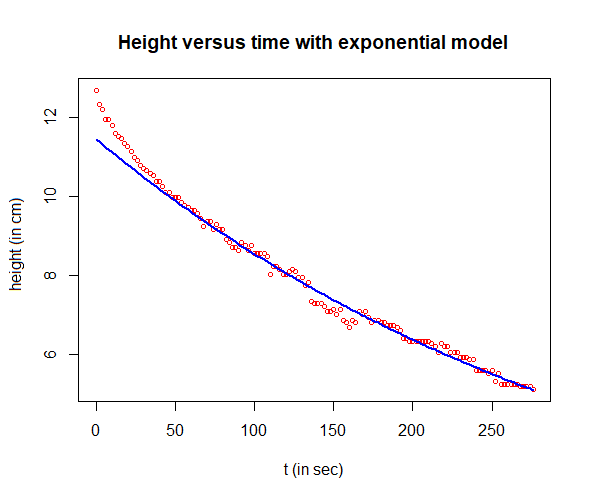
Step 3: plotting the residuals
The regression curve initially does not accurately describe the measurement data, but that is not the most important aspect. What matters is that the residuals once again exhibit an exponential decay pattern. The R script below illustrates this.
# compute residuals residuen and plot them
residuals <- H - H_fit
plot(t, residuals, type="p", pch=16, cex=0.7, col="red",
xlab="t (in sec)", ylab="residual (in cm)",
main="residuals versus time")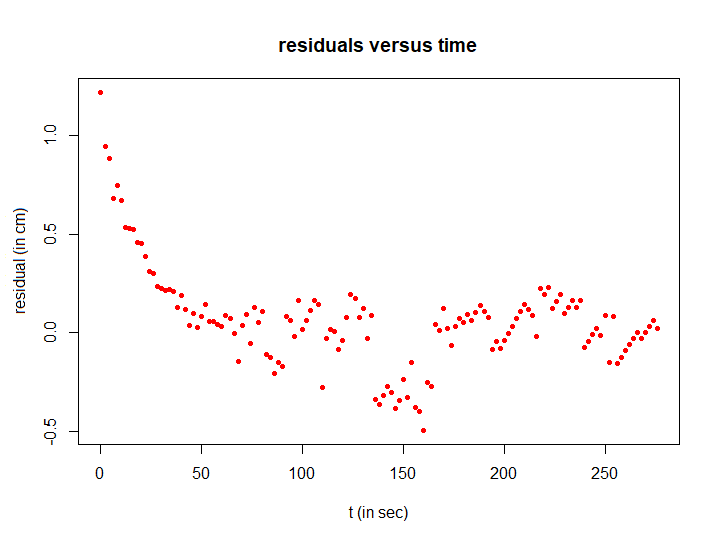
Step 4: describing the first part of measured data with exponential regression
We can calculate an exponential model for the first part of the residuals in the same way as before. For a change, let's perform a nonlinear fit with an estimate of initial parameter values using the method of partial sums. The R script below provides the exponential function fit of the residuals and a corresponding plot..
# select measured data in the first 60 seconds
dataselection <- c(t<60)
ts <- t[dataselection]
Rs <- residuals[dataselection]
# do a parameter estimation via partial sums
# by the way, logarithmic transformation works as well
N <- length(ts)/2
Nplus1 <- N+1
dt <- 2
s1 <- sum(Rs[1:N])
s2 <- sum(Rs[Nplus1:30])
r <- (log(s2)-log(s1)) / (N*dt)
h <- exp(r*dt)
a <- s1 * (1-h) / (1-h^N)
# apply nonlinear regression
nlfit <- nls(formula = Rs ~ a_2*exp(r_2*ts), start = list(a_2=a, r_2=r))
coeffs <- coefficients(nlfit)
a2 <- coeff[1]
r2 <- coeff[2]
R_fit <- a2*exp(r2*t) # teken residuen in een scatterplot en voeg expontiële fit toe
plot(t, residuals, type="p", pch=16, cex=0.7, col="red",
xlab="t (in sec)", ylab="residual (in cm)",
main="residuals versus time with exponential model")
lines(t, R_fit, type="l", col="blue", lwd=2) 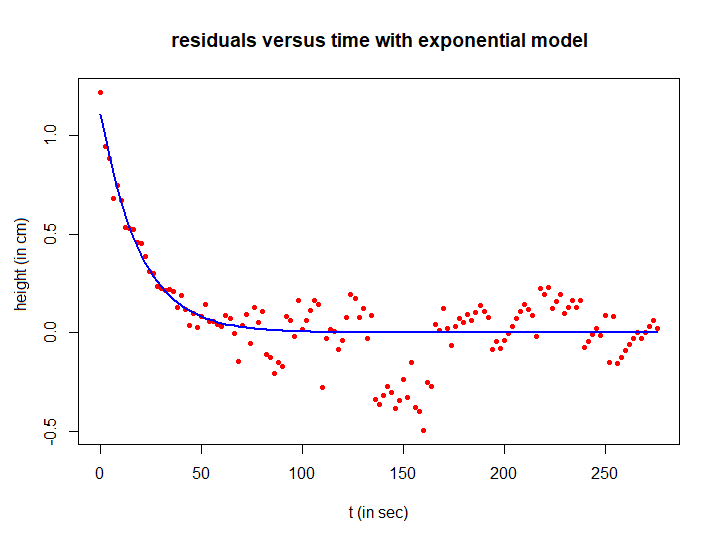
Step 5: Putting all together
If we add both calculated regression curves, we obtain a nonlinear regression of the measured height of the beer foam over time:
# final result
plot(t, H, type="p", pch=1, cex=0.7, col="red",
xlab="t (in sec)", ylab="height (in cm)",
main="Height of beer foam versus time plus bi-exponential model")
lines(t, H_fit + R_fit, type="l", col="blue", lwd=2)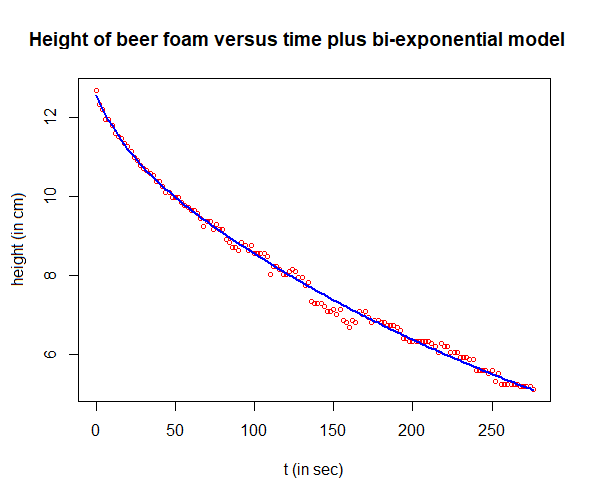
Step 6: Nonlinear regression with the final model and parameter estimates already found
It is even better to use the found parameter values as starting values for the following regression model \[\text{hoogte} = a_1\cdot e^{r_1\cdot t}+ a_2\cdot e^{r_1\cdot t}\] The R script below does the job.
# final nonlinear regression:
# apply bi-exponential regression
nlfit <- nls(formula = H ~ a_1*exp(r_1*t) + a_2*exp(r_2*t),
start = list(a_1=a1, r_1=r1, a_2=a2, r_2=r2))
print(summary(nlfit)) # print results
# plot the mmeasured data and the regression curve
plot(t, H, type="p", pch=1, cex=0.7, col="red",
xlab="t (in sec)", ylab="height (in cm)",
main="Height of beer foam versus time plus bi-exponential model")
lines(t, predict(nlfit), type="l", col="blue", lwd=2)
# compute the correlation between measured data and modelling result
cat("correlation coefficient =", cor(H,predict(nlfit)))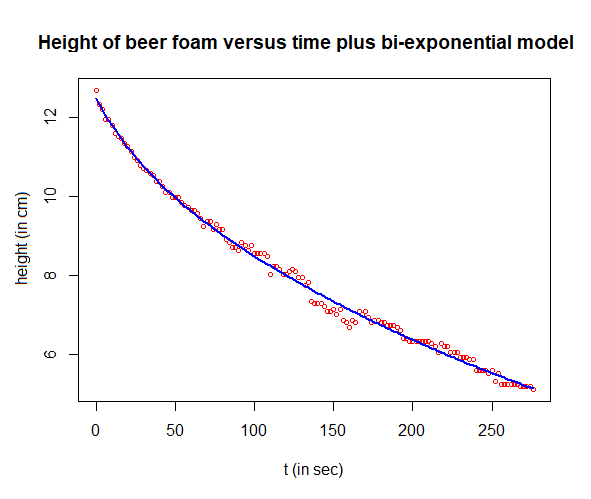
The final parameter estimates are tabulated below. The correlation coefficient between the measurement data and model results has also been calculated and printed. The end result is of high quality!
Formula: H ~ a_1 * exp(r_1 * t) + a_2 * exp(r_2 * t)
Parameters:
Estimate Std. Error t value Pr(>|t|)
a_1 1.121e+01 1.119e-01 100.191 < 2e-16 ***
r_1 -2.822e-03 5.164e-05 -54.641 < 2e-16 ***
a_2 1.284e+00 1.087e-01 11.815 < 2e-16 ***
r_2 -3.338e-02 5.665e-03 -5.892 2.89e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.1399 on 135 degrees of freedom
Number of iterations to convergence: 8
Achieved convergence tolerance: 7.89e-06
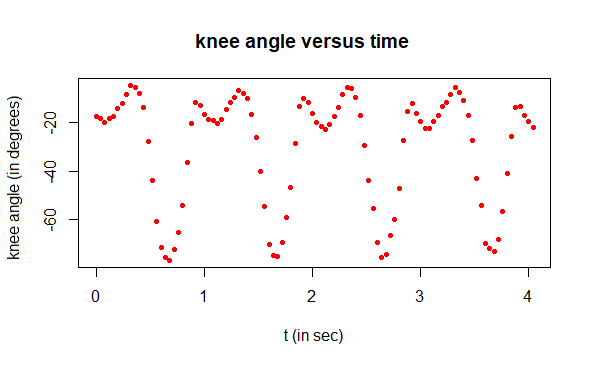
correlation coefficient = 0.9975641Knee angle during walking: harmonic analysis A girl's knee angle was measured while walking at a constant speed on a treadmill. The claim is that this angle can be describedwell as the sum of two sinusoids. This is an example of harmonic analysis. This approximation can be determined as follows. The R script is executed in steps.
Step 1: reading in and plotting measured data
# read data from csv file kneedata <- read.csv("kneeanglemeasurement.csv", header = TRUE, sep = ";", dec = ".", na.strings="", fill = TRUE) t <- kneedata$time # time in sec angle <- kneedata$angle # knee angle in degrees # plot knee angle data in a scatter plot plot(t, angle, type="p", pch=16, cex=0.7, col="red", xlab="t (in sec)", ylab="knee angle (in degrees)", main="knee angle versus time")
Step 2: describing measured data with sinusoidal regression
This resembles a periodic motion with a frequency of about 1 s-1 . Let's first approach the measured data with a simple sinusoidal regression model \[\text{angle} = a_1\sin(b_1t+c_1)+d_1\] where the sine function takes arguments in degrees. For this we need to define a sine function in degrees in R. We estimate the starting values of the parameters for a nonlinear regression method as follows:
- from the frequency follows an estimate for the starting value of \(b_1\), namely: \(b_1=360\);
- we get the amplitude \(a_1\) as half the difference between extreme angular values;
- we estimate the vertical shift \(d_1\) of the sinusoid as the mean angular value because the time interval appears to be four periods;
- we initially estimate the phase angle to be equal to 0, for lack of a better answer.
The R script below produces the non-linear function fit and a matching diagram.
# define sine function in degrees instead of radians as input
sin_in_degrees <- function(degrees) {
radians <- degrees*(pi/180)
return(sin(radians))
}
# apply sinusoidal regression (with actually two components)
fit1 <- nls( formula = angle ~ a1*sin_in_degrees(b1*t+c1) + d1,
start=list(a1=(max(angle)-min(angle))/2,
b1=360, c1 = 0, d1=mean(angle)))
coeffs1 <- coefficients(fit1)
a1 <- coeffs1[1]
b1 <- coeffs1[2]
c1 <- coeffs1[3]
d1 <- coeffs1[4]
cat("a1 =", round(a1,2), "b2 =", round(b1,2),
"c1 =", round(c1,2), "d1 =", round(d1,2), "\n")
angle1 <- a1*sin_in_degrees(b1*t+c1) + d1
# plot measured data and regression curve
plot(t, angle, type="p", pch=16, cex=0.7, col="red",
xlab="t (in sec)", ylab="knee angle (in degrees)",
main="knee angle versus time plus sinusoid")
lines(t, angle1, type="l", col="blue", lwd=2)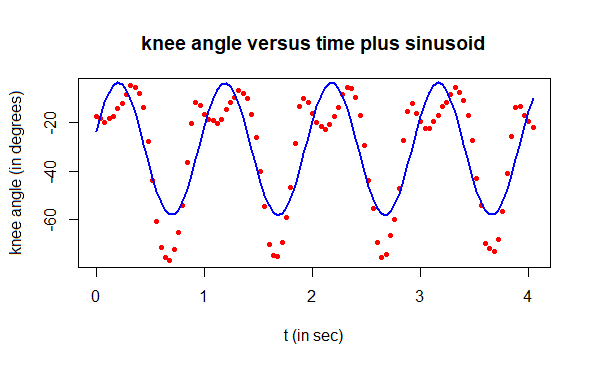
Step 3: plotting of residuals
The regression curve does not describe the measured data accurately, but that is not so important. What matters is that the residuals exhibit a periodic character once again. The R script below demonstrates this.
# compute residuals and plot them
residuals <- angle - angle1
plot(t, residuals, type="p", pch=16, cex=0.7, col="red",
xlab="t (in sec)", ylab="residual (in degrees)",
main="residuals versus time")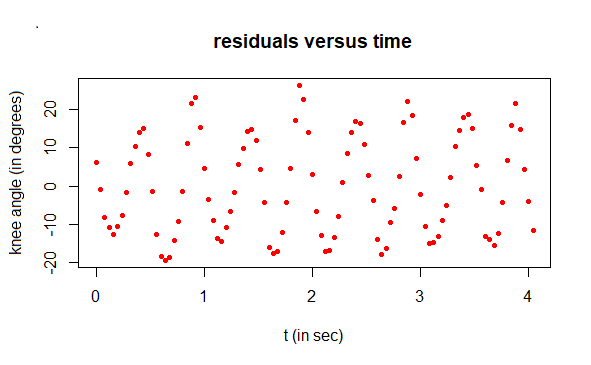
Step 4: describing of residuals with again sinusoidal regression
This appears to be a periodic motion with a frequency of approximately 2 s-1. First, we'll approximate the residuals with a sinusoidal regression model \[\text{hoek} = a_2\sin(b_2t+c_2)+d_2\] where the sine function takes arguments in degrees. We estimate the initial values of the parameters for a nonlinear regression method in the following way:
- from the frequency, we obtain an estimate for the initial value of \(b_2\), namely: \(b_1=720\);
- the amplitude \(a_2\) is obtained as half of the difference between the extreme values of residuals;
- the vertical shift \(d_1\) of the sinusoid is estimated as close to 0. So we take \(d_2=0\);
- Looking at the graph, it appears to be the negative of a sine. This observation leads us to estimate \(c_2=180\) (corresponding with \(\pi\)).
The R script below gives the nonlinear function fit of the residuals and a corresponding diagram.
# apply sinusoidal regression (with actually two components)
fit2 <- nls( formula = residuals ~ a2*sin_in_degrees(b2*t+c2) + d2,
start = list(a2=(max(residuals)-min(residuals))/2,
b2=720, c2=180, d2=0))
coeffs2 <- coefficients(fit2)
a2 <- coeffs2[1]
b2 <- coeffs2[2]
c2 <- coeffs2[3]
d2 <- coeffs2[4]
cat("a2 =", round(a2,2), "b2 =", round(b2,2),
"c2 =", round(c2,2), "d2 =", round(d2,2), "\n")
angle2 <- a2*sin_in_degrees(b2*t+c2) + d2
# plot residuals and regression curve
plot(t, residuals, type="p", pch=16, cex=0.7, col="red",
xlab="t (in sec)", ylab="residual (in degrees)",
main="residuals versus time plus sinusoid")
lines(t, angle2, type="l", col="blue", lwd=2)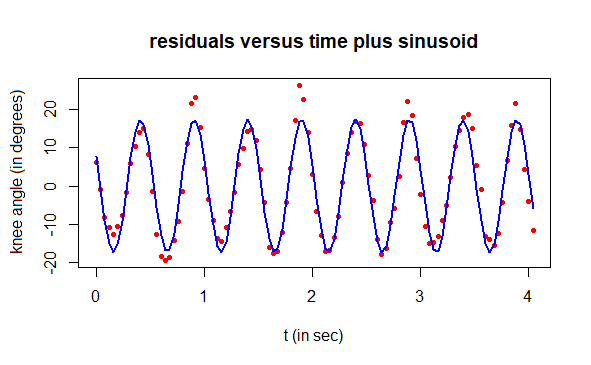
Step 5: putting it all together
If we add the two calculated regression curves, we have a nonlinear regression of the measured knee angles during walking:
# final result
plot(t, angle, type="p", pch=16, cex=0.7, col="red", ylim=c(-80,0),
xlab="t (in sec)", ylab="knee angle (in degrees)",
main="knee angle versus time plus regression curve")
lines(t, angle1 + angle2, type="l", col="blue", lwd=2)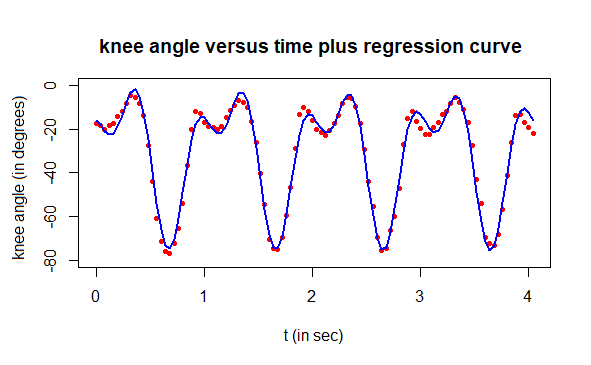
Step 6: Nonlinear regression with final model and parameter estimates already found
It is even better to use the parameter values found so for as starting values for the next regression model \[\text{height} = a_1\sin(b_1t+c_1)+a_2\sin(b_2t+c_2)+d\] The R script below does the job.
# ultimate nonlinear regression:
# apply sinusoidal regressie with twee sine functions
nlfit <- nls(formula = angle ~
a_1*sin_in_degrees(b_1*t+c_1) +
a_2*sin_in_degrees(b_2*t+c_2) +
d,
start = list(a_1=a1, b_1=b1, c_1=c1,
a_2=a2, b_2=b2, c_2=c2,
d=d1))
print(summary(nlfit)) # print results
# plot the measured data and the regression curve
plot(t, angle, type="p", pch=16, cex=0.7, col="red", ylim=c(-80,0),
xlab="t (in sec)", ylab="knee angle (in degrees)",
main="knee angle versus time plus regression curve")
lines(t, predict(nlfit), type="l", col="blue", lwd=2)
# compute the correlation between data and modelling results
cat("correlation coefficient =", cor(angle,predict(nlfit)))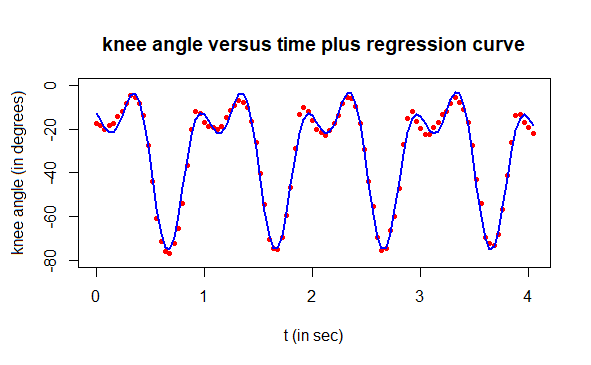
The final parameter estimates are tabulated below. Additionally, the correlation coefficient between measurement data and modelling results has been calculated and printed. The end result is of high quality!
Formula: angle ~ a_1 * sin_in_degrees(b_1 * t + c_1) + a_2 * sin_in_degrees(b_2 * t + c_2) + d
Parameters:
Estimate Std. Error t value Pr(>|t|)
a_1 27.2705 0.4548 59.96 <2e-16 ***
b_1 361.0197 0.8135 443.76 <2e-16 ***
c_1 23.1223 1.9446 11.89 <2e-16 ***
a_2 17.7445 0.4588 38.68 <2e-16 ***
b_2 723.3587 1.2204 592.72 <2e-16 ***
c_2 155.6522 2.8692 54.25 <2e-16 ***
d -30.7267 0.3206 -95.83 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 3.22 on 95 degrees of freedom
Number of iterations to convergence: 4
Achieved convergence tolerance: 5.11e-06
correlation coefficient = 0.9908715